
Microarrays and Microdeletions: Key Concepts Summarized
WHAT IS IT?
A microarray describes a newer technology that can identify small duplications or deletions of genetic material that previously could not be identified using conventional karyotyping alone. It has become a critical tool to help identify submicroscopic chromosomal deletions/duplications that underlie clinically significant syndromes in the prenatal period and throughout the lifespan.
Key Concepts
What is a Deletion?
- A deletion describes a chromosomal break where genetic material is lost
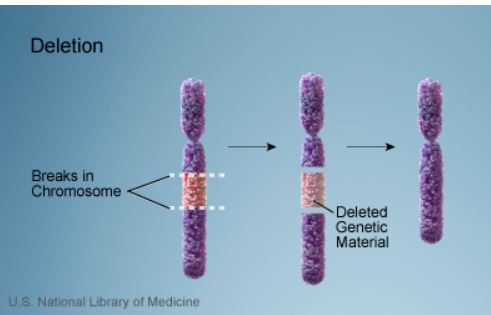
Credit: US National Library of Medicine
- Deletions can be large or small, and can occur anywhere along a chromosome
- Terminal deletion (end of a chromosome)
- Interstitial (within the chromosome)
- Duplication describes when additional material is gained
What is a Microdeletion?
- Definition is based on size of missing DNA sequence
- 1Mb (megabase) = 1 million base pairs
- Conventional karyotype is based on light microscopy and can usually only detect deletions > 5 Mb
- Microdeletions refer to deletions smaller than those that can be seen on karyotype – i.e., < 5 Mb
What is a CNV (copy number variant)?
- Deletions or duplications are ≥ 1 kb (1000 base pairs) to many hundreds of Kb in size
- Small CNVs are common and found in 5 to 10% of individuals in the general population
- Only 1 to 2% will have CNVs > 1Mb
- Most CNVs are simply a part of normal human variation
- If CNVs delete or add sufficiently large number of genes or result in a breakpoint within a gene, they may have significant clinical consequences
Key Points:
CGH vs SNP arrays
CGH (Comparative genome hybridization)
- The array is constructed with single short DNA sequences from the regions of interest
- ‘Test’ and ‘control’ DNA are labelled with different fluorescent colors and then hybridized (annealed) to complementary sequences of interest on the array
- The relative amounts of test vs control DNA can be measured
- Excess control DNA signal color signifies a deletion in the test sample
- Excess test DNA signal color signifies duplication in the test sample
SNP array
- The array is constructed with short DNA sequences
- Each region of interest will have two DNA sequences, representing the two possible alleles (versions) of known SNPs
- Only ‘test DNA’ is labelled and hybridized to the allele specific probes within the array
- Relative intensity of the hybridization signal is used to detect if DNA sequence is deleted or duplicated
- Low/absent intensity signal signifies deletion
- Excess signal signifies duplication
Additional capabilities of SNP array compared to CGH
- Can detect triploidy, maternal cell contamination and mosacism
- Can detect consanguinity and uniparental disomy (UPD) which can be associated with genetic syndromes
- Rather than having one copy of each SNP allele, a region of identical SNPs, known as absence of heterozygosity (AOH), will be apparent
- Labs may report AOH, raising concern for
- Autosomal recessive disorders related to genes in that particular sequence or
- UPD, depending on chromosome and region
Learn More – Primary Sources:
ACOG Committee Opinion 682: Microarrays and Next-Generation Sequencing Technology: The Use of Advanced Genetic Diagnostic Tools in Obstetrics and Gynecology
The use of chromosomal microarray for prenatal diagnosis
ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013
Yield of additional genetic testing after chromosomal microarray for diagnosis of neurodevelopmental disability and congenital anomalies: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG)
Practice guideline: joint CCMG-SOGC recommendations for the use of chromosomal microarray analysis for prenatal diagnosis and assessment of fetal loss in Canada

The Use of Microarrays in the Prenatal and Postnatal Setting – Current Professional Guidelines
CLINICAL ACTIONS:
Chromosomal microarray (also known as CMA) technology has revolutionized the practice of genetics and is now commonly ordered prenatally and throughout the lifespan. Structural chromosomal anomalies, once too small to be ‘seen’ using conventional karyotyping, can now be detected using molecular techniques. Microarrays can detect copy number variants (CNVs) and therefore identify chromosomal regions where there is excess DNA (a duplication) or missing DNA (a deletion).
Consider the following when counseling:
Benefits of Microarrays
Prenatal detection of clinically significant CNVs that would have been missed using conventional karyotyping alone
- 6% of abnormal fetuses with a normal karyotype may have pathogenic CNVs or likely pathogenic CNVs
- Isolated finding: 5.6%
- Multiple anomalies: 9.1%
- 1.7% of normal fetuses and normal karyotype may have pathogenic CNVs or likely pathogenic CNVs
Postnatal detection of significant CNVs that would have been missed using conventional karyotyping alone
- Developmental delay and intellectual disability
- An additional 12.2% – 19% pathogenic anomalies May be detected with the addition of microarray
Can be performed on tissue that is no longer viable
- If DNA is present and of sufficient quality, test can be run on stillbirth specimens or products of conception
Limitations of Microarrays
Balanced translocations
- Balanced translocations occur in approximately 1/500 individuals and are usually benign
- Because there is minimal/no additional or deleted genetic material, balanced translocations will go undetected with microarray but depending on size, may be seen with conventional karyotyping
- Serious consequences are still possible with balanced translocations due to breakpoints disrupting genes
- ACMG (2018) notes the importance of balanced rearrangements (e.g., translocation, inversion, insertion) and states that “recurrence risk counseling is indicated for offspring of parent with a balanced rearrangement
- Estimated additional diagnostic yield of 0.78 to 1.3% if a G-banded karyotype is performed following a negative microarray
Single gene disorders
- May be caused by a single or small base pair change and will not be detected on microarray
Variants of uncertain significance (VUS)
- Can occur in 1 to 2% of cases
- May cause parental anxiety and will require additional expert counseling and follow-up
- Over time, VUSs are being categorized as benign or pathogenic as additional reports are incorporated into databases
SYNOPSIS:
The data from a major NICHD study in 2012 provided the support for introducing microarray technology into prenatal clinical care, with previous studies highlighting the benefits in the postnatal period and beyond. Similar to a conventional karyotype, microarrays can detect aneuplodies and larger structural chromosomal changes. In addition, microarray technology allows for the identification of small duplications and microdeletions that would otherwise go unreported. There are limitations and expert counseling is required to provide optimal care and informed decision making.
KEY POINTS:
Prenatal Recommendations
SMFM
- Fetal structural anomalies on prenatal ultrasound or stillbirth
- Microarray replaces conventional karyotype (1A)
- Patient undergoing invasive testing and no anomalies identified
- Both options, conventional karyotype and microarray, should be discussed (1B)
SOGC & CCMG
- Microarray should be offered following a normal rapid aneuploidy screen when
- Multiple fetal malformations are detected (II-1A) or
- NT ≥3.5 mm (II-2B)
Postnatal Recommendations
ACMG
- Microarray is a first-line test in the initial postnatal evaluation in the following clinical scenarios
- Multiple anomalies not specific to a well-delineated genetic syndrome
- Apparently nonsyndromic developmental delay/intellectual disability
- Autism spectrum disorders
- There may be a role in other less well-studied indications and further study is recommended
NOTE: Professional bodies universally recommend expert pre- and post-test counseling whenever microarray is being considered
Want to hear about the latest clinical summaries?
Get Your Free Newsletter »
Learn More – Primary Sources:
ACOG/ SMFM Committee Opinion 682: Microarrays and Next-Generation Sequencing Technology: The Use of Advanced Genetic Diagnostic Tools in Obstetrics and Gynecology
SMFM: The use of chromosomal microarray for prenatal diagnosis
ACMG Standards and Guidelines for constitutional cytogenomic microarray analysis, including postnatal and prenatal applications: revision 2013
Yield of additional genetic testing after chromosomal microarray for diagnosis of neurodevelopmental disability and congenital anomalies: a clinical practice resource of the American College of Medical Genetics and Genomics (ACMG 2018)
Practice guideline: joint CCMG-SOGC recommendations for the use of chromosomal microarray analysis for prenatal diagnosis and assessment of fetal loss in Canada
NEJM (2012): Chromosomal Microarray versus Karyotyping for Prenatal Diagnosis

NIPS vs. Microarray for Pathogenic Results
PURPOSE:
This study by Sotiriadis et al. (Prenatal Diagnosis, 2017) sought to calculate the proportion of pathogenic results that would be picked up by array comparative genomic hybridization (aCGH) compared to NIPS.
METHODS:
Comparative Retrospective Study
RESULTS:
This study included 2,779 fetuses that underwent invasive prenatal diagnosis using aCGH, with indications including large NT, standard 1st and 2nd trimester screening, fetal anomalies, maternal age, personal or family history of genetic issues, patient request or other (e.g. infection). Patients who were referred for screen positive NIPS were excluded. The investigators assumed a simulated NIPS panel comprised of common aneuploidies, trisomies 21, 18, 13, but also sex chromosome aneuploidies including monosomy X, 47, XXX, 47, XYY, and 47, XXY. NIPS would detect 28.0% (95% CI 14.3-47.6) of detectable aCGH pathogenic results for NT between 95th to 99th centile, 14.3% (95% 5.0-34.6) for NT > 99th centile, 34.2% (95% CI 21.1-50.1) for high-risk first-trimester results, 52.4% (95% CI 32.4-71.7) for second-trimester markers and 50.0% (95% CI 26.8-73.2) for advanced maternal age. Overall, the rate of aCGH pathogenic/likely pathogenic results was 5.0% of which 44.0% (95% CI 36.0-52.2) would have been missed by NIPS alone. This paper yet again reinforces the fact that NIPS is a screening test only and, particularly in the age of aCGH, approximately half of pathogenic findings detectable with invasive testing will be missed.
Learn More – Primary Sources:
Non-invasive prenatal screening versus prenatal diagnosis by array comparative genomic hybridization: a comparative retrospective study

Should Amniocentesis or Chorionic Villus Sampling Be Offered to All Pregnant Women?
CLINICAL ACTIONS:
Invasive prenatal diagnostic testing usually refers to amniocentesis (analysis of amniotic fluid cells) or chorionic villus sampling (placental cells). Prenatal healthcare providers should
- Offer all patients the option of prenatal invasive testing or prenatal screening
- Counsel patients that unlike invasive tests which cover all chromosomal abnormalities, traditional screening tests or even the newer cfDNA (NIPS / NIPT) tests will only screen for specific and limited chromosomal abnormalities
- although cfDNA has superior detection for Down syndrome, traditional first-trimester combined screening can detect additional structural and anatomical anomalies because of the ultrasound component of the test
- Identify the following risk factors for fetal aneuploidy and consider referral to genetic counseling and high risk OB services if patient requires more in depth counseling or has additional concerns
- Maternal age of 35 or older at EDD: the risk for chromosomal aneuploidy increases throughout the reproductive lifespan, not just after age 35
- If either parent of the fetus has an unusual chromosome makeup or aneuploidy, such as an additional X or Y chromosome
- ACOG states
A patient’s baseline risk for chromosomal abnormalities should not limit testing options; serum screening with or without NT ultrasound or cell-free DNA screening and diagnostic testing (CVS or amniocentesis) should be discussed and offered to all patients regardless of maternal age or risk for chromosomal abnormality
Allowing patients to select diagnostic or screening approaches for the detection of fetal aneuploidy and/or genomic changes that are consistent with their personal goals and preferences
Informing all pregnant women that diagnostic testing (CVS or amniocentesis) is an option for the detection of chromosome abnormalities and clinically significant CNVs
SYNOPSIS:
The cells obtained from amniocentesis or CVS are analyzed to determine if the number of chromosomes are correct (46) and whether there are structural changes such as deletions or duplications. Routine karyotyping is done using light microscopy. If changes are smaller than the resolution of a microscope, then molecular techniques are required and these small alterations are called microduplications or microdeletions (see ‘Related ObG Topics’ below). Presently, despite major advances in screening technologies, diagnosis of fetal aneuploidy still requires an invasive test.
KEY POINTS:
Miscarriage Risks
- In expert hands, there is a 0.1 to 0.3% chance of miscarriage associated with invasive prenatal testing
- Cochrane Review (Alfirevic et al., 2017) has released its review on amnio/CVS safety
- 2nd trimester amnio increased risk of pregnancy loss, but it was not possible to quantify the loss rate, based on one study that is now over 30 years old
- Early amnio (11w0d-12w6d) is associated with pregnancy loss and clubfoot compared to 2nd trimester amnio (15w0d-16w6d)
- Transcervical CVS may be associated with higher loss rate compared to 2nd trimester amnio but the quality of the evidence was downgraded due to heterogeneity between studies
- Wulff et al. (Ultrasound Obstet Gynecol, 2016)
- Using propensity scoring on a nationwide database of approximately 150,000 women, did not find an increased risk of miscarriage or stillbirth due to amnio or CVS when indications for the procedures were taken in to account (see Related OBG Topics below for review of this paper and other recent papers on this subject of procedure related fetal loss)
- Salomon et al. (Ultrasound Obstet Gynecol, 2019)
- Estimated procedure-related risk of miscarriage after amniocentesis and chorionic villus sampling (CVS)
- Performed a systematic review and meta-analysis, covering 20 controlled studies
- The authors concluded
…amniocentesis is associated with a procedure-related risk of 1:300 at most, or more likely, no significant increase in risk
With regard to CVS, our results demonstrate that, there is no significant procedure-related risk associated with undertaking this procedure
Routine Karyotyping or Microarray?
- Abnormal prenatal ultrasound with structural abnormality
- A chromosomal microarray analysis that can detect submicroscopic changes is recommended
- Standard karyotype may miss 6% of important chromosome changes
- Normal prenatal ultrasound
- A chromosomal microarray can be offered because 1.7% of significant chromosome changes will not be detected on a standard karyotype approach
- Microarray limitations to discuss with patients (see ‘Related ObG Topics’ below for more on the benefits and limitations of microarray analysis)
- In a small number of cases, the laboratory may identify copy number variants of uncertain significance (VUS), also referred to as variants of uncertain significance (VOUS)
- Over time, as databases grow, VUSs can be re-categorized as benign or pathogenic
- Microarrays cannot detect low levels of mosaicism (more than one cell line) or balanced translocations
- Small risk that while overall DNA appears balanced on a microarray, the breaks involved in the translocation may have disrupted a gene and lead to abnormal protein production
Additional Considerations
- A patient who would not terminate a pregnancy
- Important information that may impact the management of a pregnancy may be obtained on invasive testing beyond termination of pregnancy
- Therefore, all patients should be offered the option of screening or invasive testing and have the option of accepting or declining testing irrespective of future reproductive choices
- Diagnosis code: diagnosis code will vary depending on indication
- Procedure codes: amniocentesis- 59000; sono guidance for amniocentesis- 76946
- Procedure codes: CVS- 59015; sono guidance for CVS-76945
Learn More – Primary Sources:
ACOG Practice Bulletin No. 226: Screening for Fetal Chromosomal Abnormalities
ACOG Practice Bulletin No. 162: Prenatal Diagnostic Testing for Genetic Disorders
ACOG Committee Opinion No. 682: Microarrays and Next-Generation Sequencing Technology: The Use of Advanced Genetic Diagnostic Tools in Obstetrics and Gynecology
Amniocentesis and chorionic villus sampling for prenatal diagnosis
ACOG Statement on FDA Warning on Genetic Non-Invasive Prenatal Screening Tests | ACOG
Locate a Genetic Counselor or Genetics services:
Genetic Services Locator-ACMG
Genetic Services Locator-NSGC
Genetic Services Locator-CAGC
Locate a Maternal Fetal Medicine Specialist
Maternal Fetal Medicine Specialist Locator-SMFM




